Checking in on Biochemical Kinase behaviour
Real-time measurement and understanding of kinase activity serve many purposes. It allows insights in cellular signaling and pathway elucidation but also target interactions. Recently PamGene’s technology and expertise has contributed to the development of a promising new method to validate kinases as therapeutic targets.
Nature Communications recently published a study on this chemical genetics strategy. Tom van der Wel PhD, post-doc at Leiden University’s group for Molecular Physiology, took the first steps to develop this method, taking into account the large group of so-far untargeted kinases that offer great opportunities for the development of novel molecular therapies.
Using Chemistry for a better understanding of biology
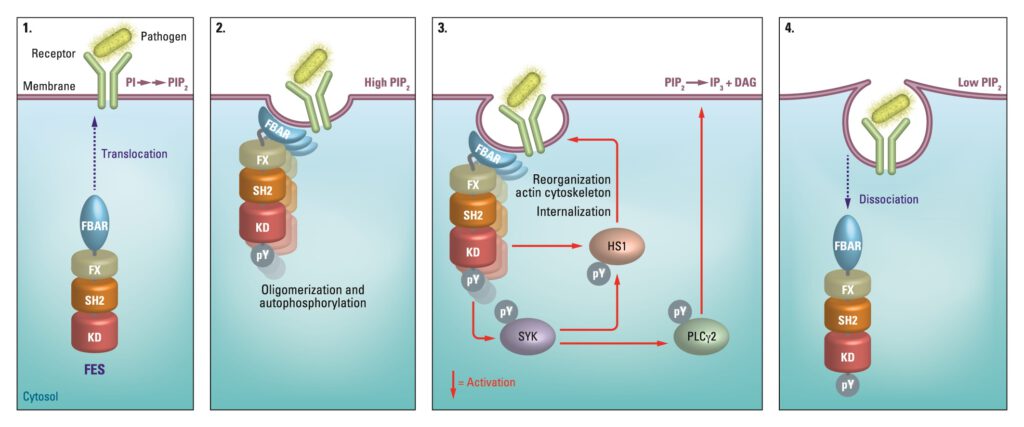
The non-receptor tyrosine kinase feline sarcoma oncogene (FES), a potential therapeutic target for cancer and immune disorders, was subject of Van der Wel’s study. “By introducing a mutation in the endogenous FES gene of HL-60 cells using CRISPR/Cas9 gene editing, we sensitize this kinase towards covalent labeling by a complementary fluorescent chemical probe. This approach allows acute inactivation of kinases with high specificity, it can be employed to study kinase function and thereby aid in its validation as therapeutic target,” tells Van der Wel.
“For our methodology, it was essential that this engineered, mutant FES kinase showed similar biochemical behavior as the wild-type kinase. This is where we got in touch with PamGene: their technology to gain insight in kinase activity and binding technology were ideally suited for this purpose.”
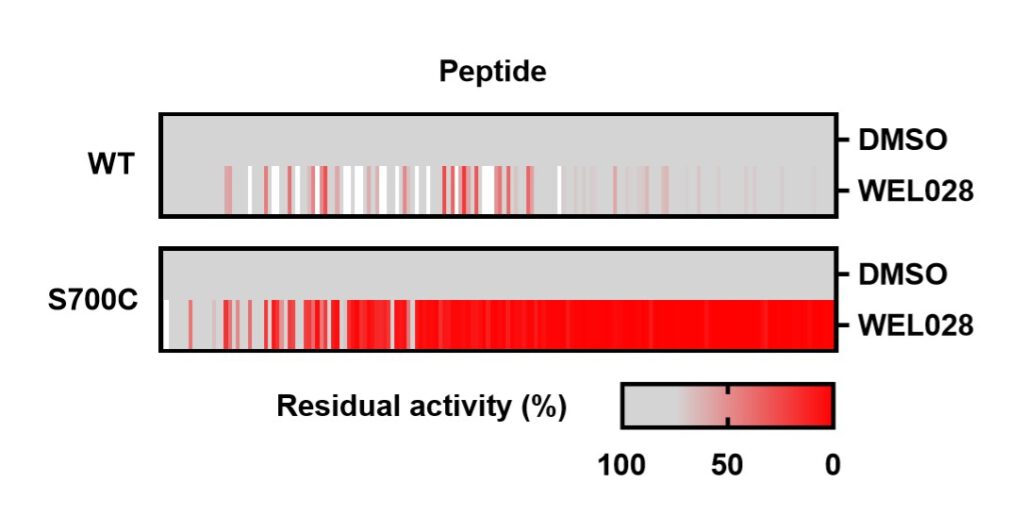
To assess whether the introduced mutation affected substrate recognition, a comparative substrate profiling assay was performed using the PamChip® microarray technology. This assay is based on the phosphorylation of immobilized peptides by purified FES and detection using a fluorescently labeled anti-phosphotyrosine antibody. Strikingly, the substrate profiles of FESWT and FESS700C were completely identical, indicating that the S700C mutation did not affect substrate recognition. A modified PamChip array to measure phosphopeptide binding to the Src homology 2 (SH2) domain of FESWT and FESS700C showed that the introduced mutation did not affect the SH2 binding profile.
“Results from PamGene’s profiling assays formed a starting point for mechanistic studies into the role of FES in neutrophils, which eventually led to the identification of a novel FES substrate in this cell type.”
Interested in learning more?
In collaboration with both Covalution Biosciences and PamGene’s kinase experts, Van der Wel published his results in Nature Communications:
https://www.nature.com/articles/s41467-020-17027-5
Would you like to learn more, leave us a message and get in touch with one of our experts.
