Are you interested in gaining new publish-worthy signal transduction insights based on real-time cell activity? Measuring kinase activity provides deeper understanding of cellular signaling allowing pathway elucidation.
Are you interested in gaining new publish-worthy signal transduction insights based on real-time cell activity? Measuring kinase activity provides deeper understanding of cellular signaling allowing pathway elucidation.
PamGene is operating on the cutting edge of next generation kinase science and high-tech to support scientists in gaining new signal transduction pathway insights by measuring real-time kinase activity in signalling cascades. Building on 15 years of experience, PamGene has developed a unique technology with unprecedented sensitivity, making real-time measurement and understanding of kinase activity possible.
Measuring kinase activity provides deeper understanding of cellular signaling allowing pathway elucidation.
PamGene is operating on the cutting edge of next generation kinase science and high-tech to support scientists in gaining new signal transduction pathway insights by measuring real-time kinase activity in signalling cascades. Building on 15 years of experience, PamGene has developed a unique technology with unprecedented sensitivity, making real-time measurement and understanding of kinase activity possible.
Measuring kinase activity provides deeper understanding of cellular signaling allowing pathway elucidation.
Thanks to our unique and proven technology we will provide you with robust measurements, which can be applied in numerous of applications bringing your insights and research to the next level.
Thanks to our unique and proven technology we will provide you with robust measurements, which can be applied in numerous of applications bringing your insights and research to the next level.
We have over 15 years of experience in the field of kinase science and contributed to over 100+ publications with top European, North American and Japanese Institutes.
Sawada T, Hilhorst R, Rangarajan S, Yoshida M, Tanabe Y, Tamura K, Kinoshita T, Shimoyama T, van Beuningen R, Ruijtenbeek R, Tsuda H, Koizumi F.
Oncotarget. Sep 28;9(76):34229-34239. (2018)
Chocry M, Leloup L, Kovacic H.
Oncotarget. 2017 Oct 10;8(61):103710-103730.
Ferguson BD, Tan YH, Kanteti RS, Liu R, Gayed MJ, Vokes EE, Ferguson MK, Iafrate AJ, Gill PS, Salgia R.
Sci Rep. 5:10641. doi: 10.1038/srep10641. (2015)

We have over 15 years of experience in the field of kinase science and contributed to over 100+ publications with top European, North American and Japanese Institutes.
Sawada T, Hilhorst R, Rangarajan S, Yoshida M, Tanabe Y, Tamura K, Kinoshita T, Shimoyama T, van Beuningen R, Ruijtenbeek R, Tsuda H, Koizumi F.
Oncotarget. Sep 28;9(76):34229-34239. (2018)
Chocry M, Leloup L, Kovacic H.
Oncotarget. 2017 Oct 10;8(61):103710-103730.
Ferguson BD, Tan YH, Kanteti RS, Liu R, Gayed MJ, Vokes EE, Ferguson MK, Iafrate AJ, Gill PS, Salgia R.
Sci Rep. 5:10641. doi: 10.1038/srep10641. (2015)

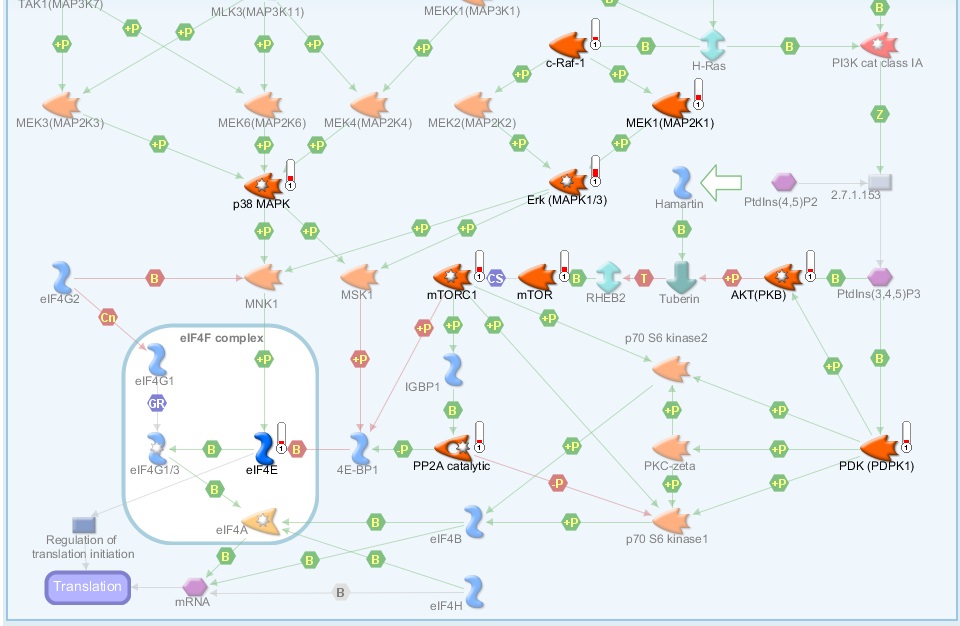
Kinases are the most intensively studied protein targets. We have developed a unique method to understand them. Our sensitive platform uses lysates obtained from only a few thousand cells or small amounts (2ug) of animal, xenografts or human tissues and cells to simultaneously determine the activity of all kinases present in these lysates.
This is accomplished by incubating the lysates across 196 to 144 tyrosine or serine/threonine kinase peptide substrates immobilized on the PamChip® microarray platform. Our 3D microarrays are spotted with peptides (i.e. phosphosites) that represent kinase targets. Kinases present in the lysates will phosphorylate the peptide substrates which are detected using fluorescently labelled antibodies. The phosphosites on the PamChips are human-derived and there is significant overlap with other organisms.
Based on current online knowledge, we compiled a comprehensive, integrated database (DB) of potential kinases that are linked to the peptides on the PamChips. This corresponds to ~350 unique kinases in literature, covering the majority of the kinome.
Kinases are the most intensively studied protein targets. We have developed a unique method to understand them. Our sensitive platform uses lysates obtained from only a few thousand cells or small amounts (2ug) of animal, xenografts or human tissues and cells to simultaneously determine the activity of all kinases present in these lysates.
This is accomplished by incubating the lysates across 196 to 144 tyrosine or serine/threonine kinase peptide substrates immobilized on the PamChip® microarray platform. Our 3D microarrays are spotted with peptides (i.e. phosphosites) that represent kinase targets. Kinases present in the lysates will phosphorylate the peptide substrates which are detected using fluorescently labelled antibodies. The phosphosites on the PamChips are human-derived and there is significant overlap with other organisms.
Based on current online knowledge, we compiled a comprehensive, integrated database (DB) of potential kinases that are linked to the peptides on the PamChips. This corresponds to ~350 unique kinases in literature, covering the majority of the kinome.

